Large repositories like The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), CPTAC, TARGET, etc., house multiomics datasets from a large number of patients. A group or cohort of patients’ datasets can be analyzed to derive insights into the disease mechanism, biomarkers identification, risk prediction, and validation for clinical research. These public repositories are hosted with the objective to aid researchers toward quality of care and treatment to the patients.
Although such large-scale studies are catalogued to study multiple ailments and diseases, there is a vast amount of data available that is centered around cancers across the world. Multiomics data like genomics, transcriptomics, epigenomics, and clinical data are largely used to track cancer progression, drug response, and disease recurrence.
Leveraging Generative AI (GenAI) for data analysis is taking the pharmaceutical and clinical industry to the next level with its speed, churning out new hypotheses based on existing knowledge. In this blog, we will discuss the power of GenAI to help users mine cohort databases using simple linguistic queries. Cohort analysis can be divided into:
- Discovery cohorts: Identify potentially new biomarkers and therapeutic targets
- Validation cohorts: Validate the findings from discovery cohorts
- Prognostic cohorts: Predict the outcome of patient cohort analysis for diseases like cancer, cardiovascular ailment, etc.
- Predictive cohorts: Predict how patients with disease will respond to treatment
Case Study: BioMedViz for Actionable insights from TCGA cohort data
TCGA is one of the widely used resources, with data from more than 12,000 patients for 33 cancer types. There are plenty of tools and web servers that provide an interface for users to interact with TCGA data and perform analysis. However, it becomes challenging when users need to perform a complex query or combine multiple datasets, either public or private.
We have developed an innovative solution, BioMedViz, leveraging GenAI for data insights from multiomics cancer cohort studies. BioMedViz reduces the need to perform data transformations, merging, and complex analysis and with a simple linguistic query can provide sophisticated and intuitive visualization of the output. This application allows users to choose the dataset and build a CSV Agent using LangChain framework that can create realistic contexts for tabular data without being trained on specific data. We have used Azure OpenAI and GPT Turbo 4K language model for agent building.
We have built an end-to-end pipeline to automate data ingestion, processing, agent building, and report generation, as seen in Figure 1.
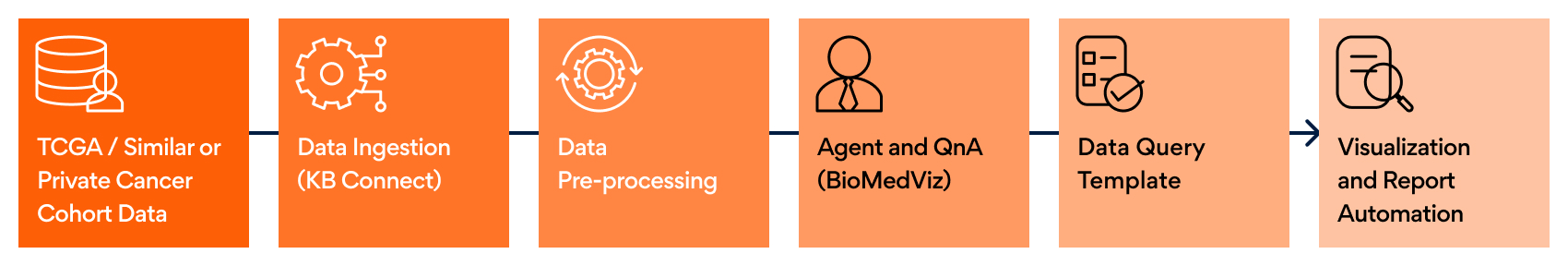
The BioMedViz pipeline has the following components:
The BioMedViz pipeline proves to be advantageous over traditional data analytics pipelines as it provides:
- End-to-end automated pipeline from cohort setup to report generation
- Support for combining multiple cohort studies
- Queries built in simple English
Figure 2 shows a sample query execution and results:
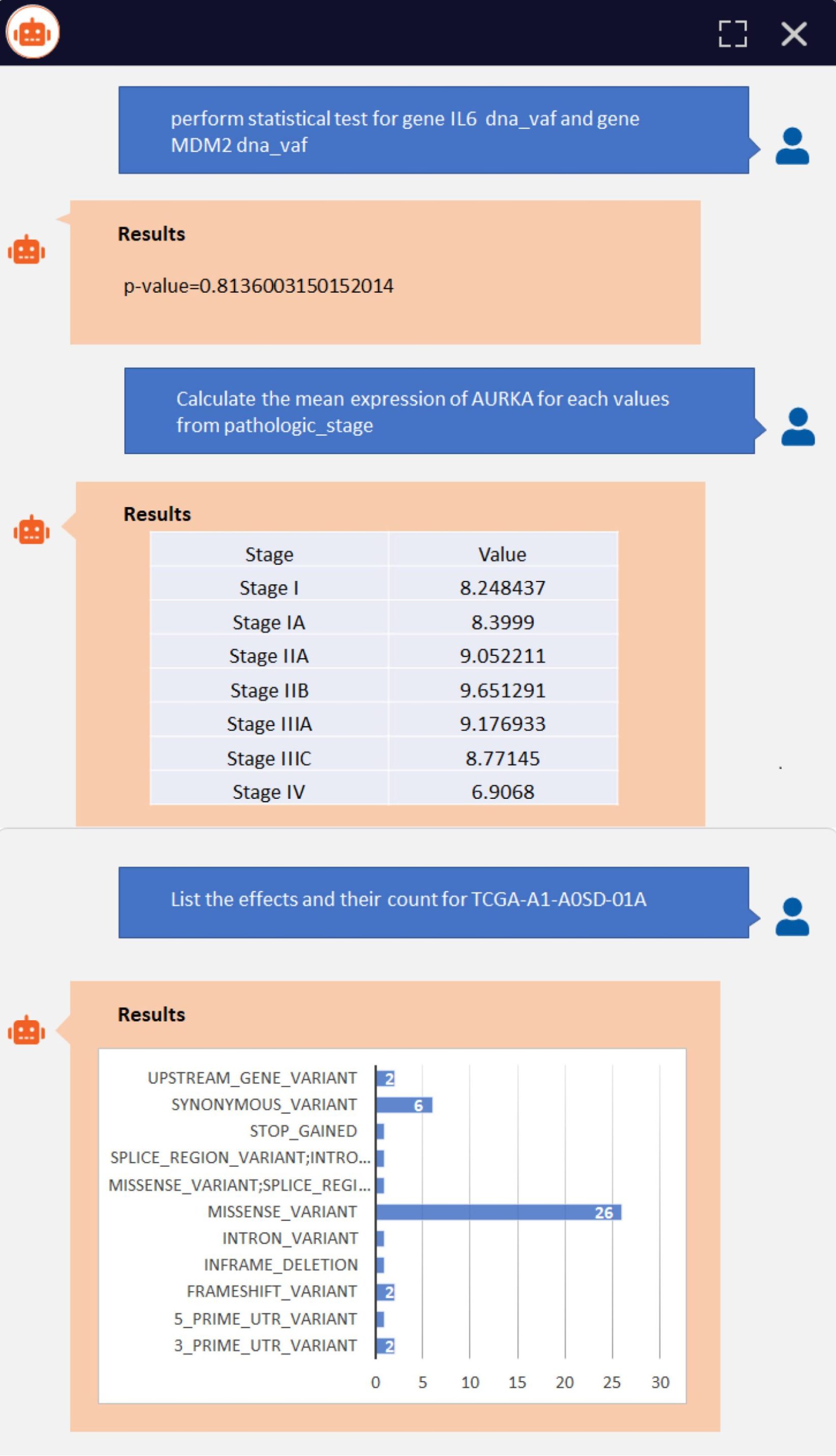
Speed and Insights from BioMedViz
BioMedViz can greatly simplify complex data analysis in a time efficient manner to detect cancer biomarkers. User can focus more on generated insights rather than data management and analytics, leading to faster results efficiently. GenAI continues to evolve, and large language models are updated often to handle larger and more relevant datasets while training. Accuracy of the data, therefore, largely depends on the underlying model and must be considered with caution. Prompt engineering-based fine tuning of models is widely used to train agents to find the most relevant results. GenAI opens up immense opportunities to perform advanced analytics on cohort multiomics data that could prove invaluable in pre-clinical, diagnostic, and therapeutic research. To learn more about BioMedViz, contact us.
Authors’ Profile
Shiva Kumar
Project Lead – Domain, Corporate CTO Organization
Indhupriya Subramanian
Project Lead – Domain, Corporate CTO Organization








