The integration of multi-modal data, encompassing genomics, transcriptomics, methylome, clinical data, including patient’s medical history of symptoms, diagnostic reports, and imaging data, has significantly transformed the landscape of personalized treatment and precision medicine. This paradigm extends to related domains like agricultural sciences and animal sciences, where multi-omics data analyses are yielding groundbreaking novel insights. This data marked by its large volume and diversity, requires seamless sharing across various user profiles. As a result, there is a need for rapid development of responsive and interactive dashboards (web apps) that can store, transform, curate, analyze and share data securely and instantly.
Over the past decade, the bioinformatics community has increasingly favored the use of “R” programming language to translate ideas into software or web applications. As an open-source programming language, R has an extensive repository of packages containing reusable code and comprehensive documentation. With the release of Shiny (an RStudio package), the development of web apps with minimum or no coding in HTML, CSS, or JavaScript is now possible. The Shiny package lets users create data analysis dashboards directly from R and Python. Currently, R and Shiny have become integral to the creation of dynamic data analysis applications, particularly, in the domains of life-sciences and healthcare.
Features of R Shiny
The features of R Shiny make it a popular tool among computational biologists and bioinformaticians. Major features of R Shiny and their advantages are listed below –
- Easy and Less Coding: In traditional web app creation, coding was required for both front-end (HTML, CSS, JavaScript) and backend (PHP, etc.). R Shiny streamlines this by handling the backend with R code and the front-end with Shiny. This reduces coding complexity and enhances efficiency.
- Reactive: R Shiny employs reactive programming to structure the logic of web app. Modifying any input values in an app, will re-execute the right part of R code, which will turn the corresponding change in the app dashboard.
- Prototyping: Shiny apps are ideal for quick web app and prototype development with minimal coding. These prototypes empower data scientists to effectively showcase the concept and functionality of their product.
- Easy to share: Shiny has made two easy ways to share the applications –
- Share as R scripts: This is the simplest way to share an app. Anyone with R and dependency packages installed on their system can run the app using the shared app script (app.R) supplemented by required materials (eg. www folder, helpers.R or styling CSS).
- Share as web page: Shiny apps can be shared/deployed as a web page. Shiny provides three ways to share apps as web page:
- Shinyapps.io: Apps can be uploaded to shinyapps.io server from R session, which provides some administration tools.
- Shiny server: It is an open-source program that can be installed on any Linux server, creating an environment to run Shiny apps. Multiple Shiny apps can be easily deployed on the same server behind firewall.
- Posit Connect: To add a layer of security, Shiny provides Posit Connect. It allows the creation of password authentication, SSL or administrator support. Shiny also provides a paid server to support performance boost and custom domains.
R Shiny Applications in Biomedical Research
Shiny is being largely used by researchers and Clinicians to visualize and interpret their data analysis in the form of web applications. It also helps showcase or share the data analysis results with others within and outside the organization. Some widely used tools that use R Shiny in genomics and multi-omics research are listed here –
- Gosling: A wrapper based on Gosling.js for interactive and scalable genomics data visualization.
- Genome viewer: Circos-based visualization of whole genome for ICGC Pancreatic cancer data.
- ShinyAIM: Interactive Manhattan plots for longitudinal genome-wide association studies (GWAS).
- GMIEC: A Shiny app which integrates multi-omics data to identify drug targets.
- ChroKit: Interactive analysis, visualization, and integration of genomic data, especially Chip-Seq and ATAC-Seq.
- ShinyFHIR: A framework to do standard-based (using HL7 FHIR) data analysis of patient data.
- ACORN: A Shiny app to generate reports on Anti-microbial Resistance (AMR) using clinical and microbiological data.
R Shiny is also widely used in the healthcare industry for efficient patient management –
- Vaccine facility simulator: Application for optimizing vaccine administration management.
- ReviewR: Application for hospital management and maintaining electronic health records (EHR).
R Shiny for Personalized Medicine using Multi-modal Data
Shiny simplifies data analysis and transformation along with the ability to visualize the data in a web application quickly and efficiently. This can be used in personalized medicine for managing data, visualizing information, and creating customized reports for handling large-scale patient healthcare data.
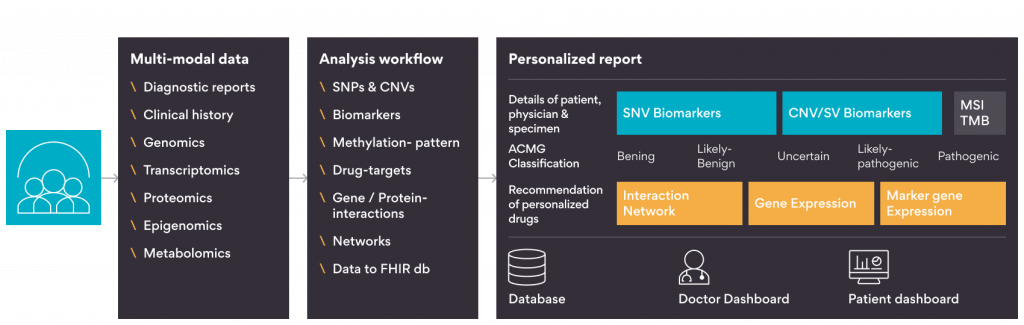
Figure 1 shows one such use case where the output of multi-modal data analysis can be fed to R Shiny apps to generate personalized reports and visualizations.
- Data collection: Patient’s clinical and genetic profile, which is multi-modal data, can be collected and saved on the cloud or on a local server.
- Multi-modal data analysis: Using customized scripts in R or Python, or readily available automated workflows like nf-core, data analysis can be done on the cloud or on a local server.
- Shiny dashboard: Shiny dashboard can consume the analyzed multi-modal data and can be used to create –
- Visualizations: The complex multi-modal data can be visualized into different plots, making data interpretation easier. Shiny supports libraries like plotly, highcharter, echart4r, etc., for visualizing reactive plots where users can change plots by changing data on the fly.
- Tertiary analysis: Tertiary or downstream analysis plays a critical role in interpreting complex data. It involves annotating diverse information about the important features of known and verified data sources. Visualizing these annotations through meaningful plots, extracts insights, thus, guiding subsequent steps. For instance, genomics-derived variants can be prioritized and ranked by clinical relevance, functional effects, and potential for therapeutic targeting.
- Reports: A concise report aids the end-user in understanding the key analysis outcomes. Sharing this report between doctors, clinicians, and patients is effortless through dedicated dashboard logins, fostering effective communication.
Limitations of Shiny
Collaborative development of R community and Rstudio team makes R and Shiny a complete package for web apps development. However, certain limitations must be considered before using these tools for development. The aesthetics of Shiny apps are difficult to change without coding in JavaScript. Shiny does not allow modification and customization of UI (User Interface) using R. There are certain incidences where Shiny apps are known to become slow and laggy, especially when handling very large datasets. R is not yet equipped to handle large datasets with the same ease as Python does. The Shiny development community is working towards improving the tool to accommodate many of the above-listed challenges.
Build R Shiny Applications with Persistent With the flexibility and ease that R Shiny offers, it can cater to data analysis requirements and development of intuitive dashboards. The R library resource at CRAN and Bioconductor provides extensive support for different biological data, which can be easily integrated in the Shiny app, thus helping the research and clinical community extensively. It is time and cost-effective and can be easily scaled for any application. There is an increasing demand for plug-and-play dashboard models, and R Shiny is capable of meeting industrial needs.
Life-sciences experts at Persistent can help build R Shiny applications on local servers or cloud-based resources. Persistent’s multi-cloud based Multi-omics solution provides integration of custom workflows, cloud operations, data management and data visualization using Shiny apps. With extensive expertise in technology and biological data, we can deliver purpose-built and tailored solutions to help the clients improve productivity. To explore more about our multi-omics solutions, reach out to us.






