During drug development , the unpredictable outcomes of the Phase II-III clinical trials (CTs) and high failure rates have been identified as the major cause for the exorbitant cost and longer timelines.1
A failed CT is a major setback for any BioPharma company due to significant loss of time, efforts, finances, and strategic focus. Therefore, BioPharma companies often undertake a thorough post-hoc CT failure analysis using all types of data accrued during drug discovery and development life cycles.2
Typically, such exercise is undertaken to:
- Investigate potential mechanisms underlying CT failure.
- Identify key scientific concepts that should be considered for improved design and implementation of future CTs to bolster the probability of success.
- Generate rational scientific hypothesis for designing experimental validation of novel therapeutic targets (for drug discovery pipelines) and putative novel biomarkers (for next-generation CT design).
Such analysis depends on big data mining of datasets generated via consented clinical meta-data from CT participants with information about genes, proteins, signaling pathways, cellular physiology, drug targets, molecular pharmacology, pharmacogenomics and their clinical phenotypes. However, given the biological complexity and high volume of such multi-disciplinary datasets, big data mining and analysis is time consuming and seldom yields meaningful and actionable insights.3 Clearly, such data mining challenge represents an increasing yet unmet need of the BioPharma industry.
To overcome these challenges, biomedical researchers are embracing in silico strategies that involve advanced computational analysis for automated integration and analysis of large datasets. In this scenario, Knowledge Graphs (KG) are becoming an increasingly popular computational tool for storing and accessing large volume of complex datasets.
KGs are graph representations where information exists in entities that are represented in the form of nodes and the relationships as edges.4 As depicted in the diagram below , these KGs serve as an alternative to traditional relational databases that are often difficult to maintain, scale up, or update with new information or out-of-date information. The power of KGs is evident in how it captures the connectivity information available in large and complex datasets, which helps to depict a better representation of data / features. The data / features can be processed using Machine Learning or Deep Learning learning algorithms to achieve state-of-the-art performance for a particular problem. At Persistent, we are actively using KGs for applications such as fraud detection, recommender systems, natural language question-answering in various disciplines.
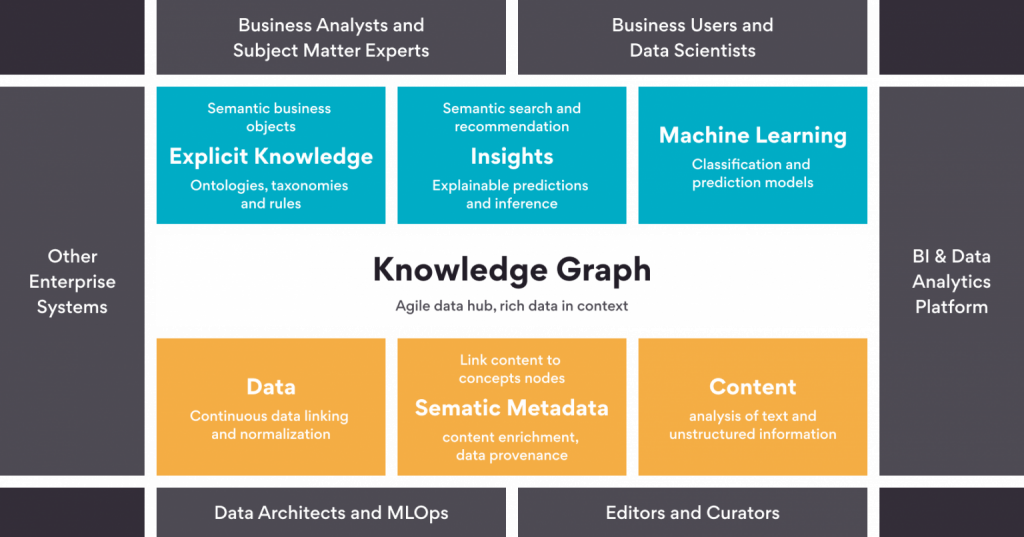
Emerging BioPharma industry trends indicate an explosion of multimodal biological datasets generated by public consortia as well as proprietary datasets. In this milieu, KG based data mining will continue to gain more uptake for pragmatic biopharmaceutical applications in near future.
Therefore, our team has recently created “OmniKG” which is a bioinsights platform for big data mining of biomedical data using a machine learning driven KG approach that reduces analysis time, human effort and subjectivity ( Figure 2).
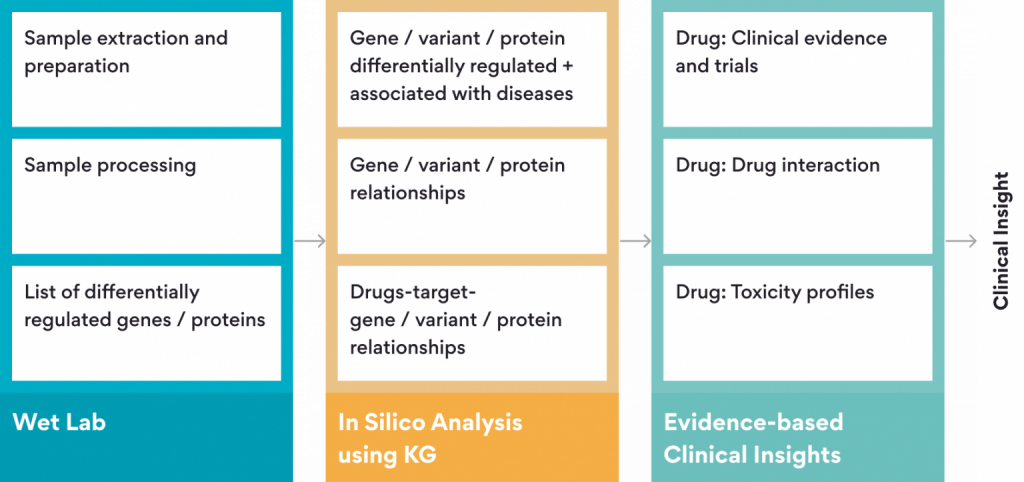
Using the OmniKG platform, we have demonstrated how KGs can be used to gain newer insights and drive innovation in areas of :
- Drug Discovery and Development– To predict and prioritize drug discovery strategies
- Preclinical Research — To identify drug-drug interactions
- Clinical Trials — To recommend drug combinations, analyze clinical trial outcomes
In the recently published white paper, we demonstrate the application of the OmniKG platform for post-hoc analysis after CT failure . IMbassador250 is a failed Phase III ImmunoOncology CT comparing atezolizumab with enzalutamide vs enzalutamide alone in patients with metastatic castration-resistant prostate cancer that was pre-maturely terminated due to severe immune toxicities.5 We have investigated plausible molecular mechanisms for identifying toxicity biomarkers associated with the drug combinations use in the CT. Using published CT data and open-source KGs ( i.e., Hetionet ; Drug Repurposing Knowledge Graphs), we deployed a dual data-mining strategy with KG embedding and deterministic approach. A six-node KG network (i.e., Drug-Gene-Disease-Pathway-Biological Process-Anatomy) was established for analysis of all potential intersecting biological pathways between enzalutamide and atezolizumab.
Our KG networks identified plausible common genes and pathways for the two drugs such as :
- Contributing gene clusters and signaling pathways
- Drug distribution pathways
- Immune-associated biomarkers
By validating our in silico KG-generated results with published literature, we were successful in generating biologically plausible hypotheses linking CT-reported clinical outcomes with molecular mechanisms revealed by our KG networks.
In near future , Persistent teams will explore applications of the OmniKG platform for BioPharma industry in the areas of Clinical Proteomics guided biomarker discovery, preclinical animal experimentation, regulatory sciences and post-marketing surveillance.
Do not miss our white paper on Knowledge Graphs for Post-Hoc Analysis of Failed Clinical Trials to get detailed insights on this topic. Reach out to us, to know more about our Healthcare & Life Sciences offerings.






