Multi-omics analysis allows the combined study of a variety of data like genomics, transcriptomics, proteomics, lipidomics, and metabolomics, to understand their interplay in a biological system. There are two main approaches which can be used to analyze multiple omics data: integrative and sequential. In an integrative approach, multiple data types are combined into one matrix using different mathematical techniques to identify biological features that play a significant role at various levels. With sequential analysis, results of one omics data analysis pipeline acts as an input to another omics analysis pipeline and so on. Sequential analysis helps in capturing the flow of information in a system.
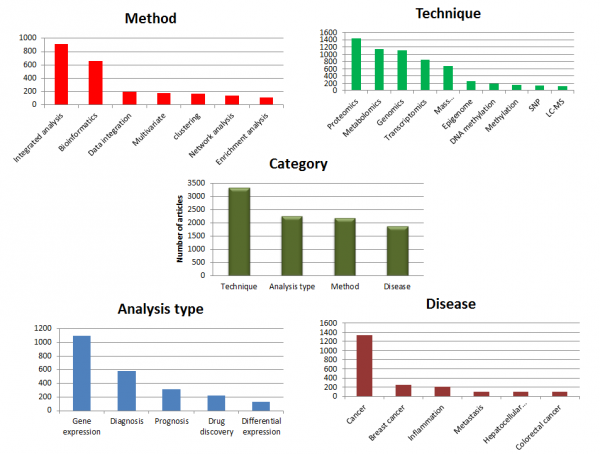
We performed a simple study to understand how multi-omics approaches are used in research. We collected and classified published information in PubMed using different search terms like multi-omics, omics, and cross-omics. The research resulted in approximately 7000 articles. We have captured current trends in the area of multi-omics by grouping these research articles based on Technique, Method, Analysis type and Disease type.
We further drill-down each category to identify what is topping in the respective categories. With regard to the “Method” category, integrated analysis is a widely used method to find novel insights into the biology of data. Within the “Analysis type” category, gene expression analysis plays a key role in understanding biological outcome or progression. For the “Technique” category, proteomics and metabolomics are preferred to better enable the understanding of the gene function and biological processes. In the “Disease” category, cancer is at the center of many studies, particularly breast cancer.
We have used BioGyan, a Java-based application to collect, annotate and rank research articles from PubMed.